Penguin Study Data Visualization & Machine Learning by using Tidymodels
In this blog post, we will visualize and carry out some machine learning models on the penguin datasets form TidyTuesday.
library(tidyverse)
library(tidymodels)
theme_set(theme_light())penguins <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-07-28/penguins.csv') %>%
filter(!is.na(bill_length_mm),
!is.na(sex)) %>%
mutate(species = factor(species))
penguins_raw <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-07-28/penguins_raw.csv') %>%
janitor::clean_names() %>%
select(-c(stage, region)) %>%
mutate(species = str_remove(species, "\\s.+"),
sex = str_to_title(sex)) Penguins' bill:
penguins %>%
ggplot(aes(bill_length_mm, bill_depth_mm, color = sex)) +
geom_point(aes(size = flipper_length_mm)) +
geom_text(aes(label = island), alpha = 0.5, vjust = 1, hjust = 1, check_overlap = T) +
facet_wrap(~species) +
scale_size_continuous(range = c(1,5), breaks = c(180, 200, 230)) +
theme(strip.text = element_text(size = 13),
plot.title = element_text(size = 16)) +
labs(x = "bill length (mm)",
y = "bill depth (mm)",
color = NULL,
size = "flipper length (mm)",
title = "Bill Length & Depth for All 3 Species",
subtitle = "Text refers to the island where penguins are located")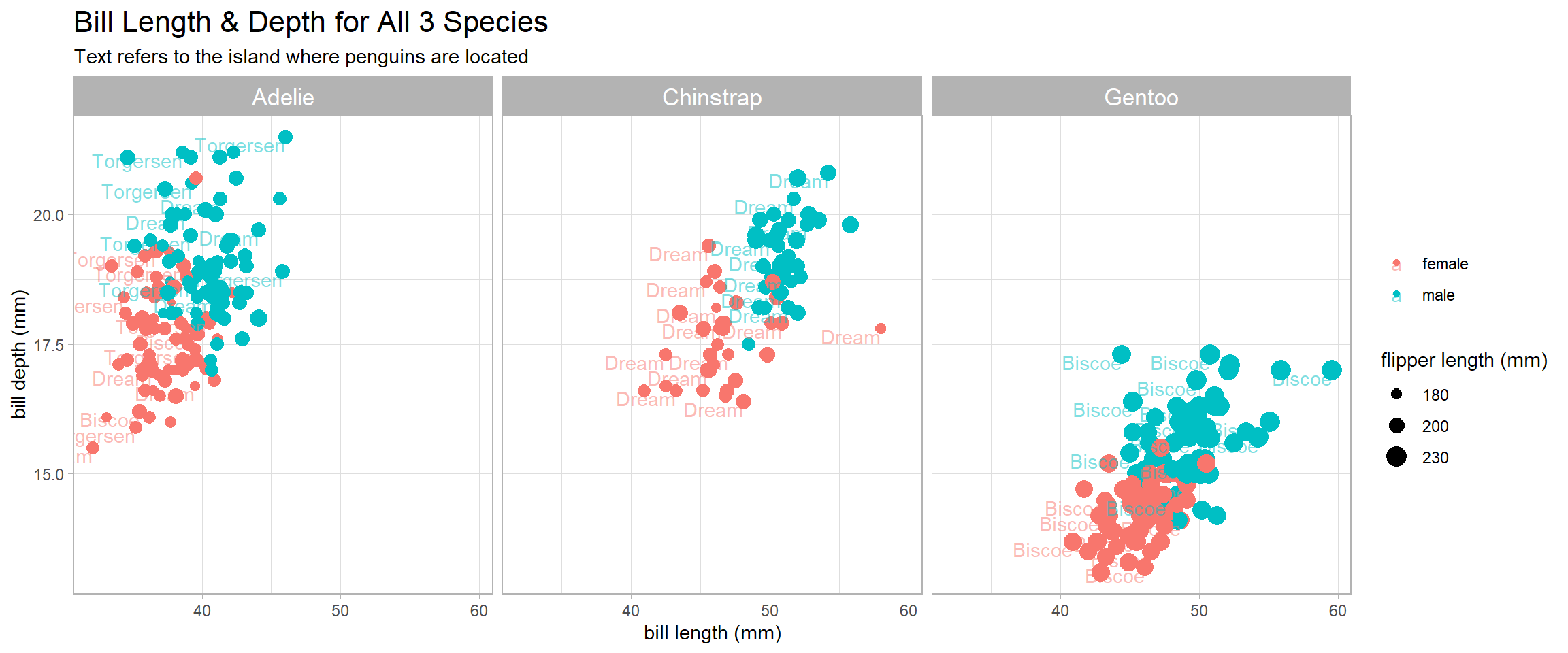
Based on the plot above, it is interesting to find out that Adelie penguins have a deeper bill than their Gentoo counterparts, yet shorter flipper and shorter length.
Penguins' body mass
penguins %>%
ggplot(aes(sex, body_mass_g, fill = species)) +
geom_boxplot() +
facet_wrap(~year) +
theme(strip.text = element_text(size = 13),
plot.title = element_text(size = 16)) +
labs(x = "",
y = "body mass (g)",
title = "Yearly Body Mass across All Species")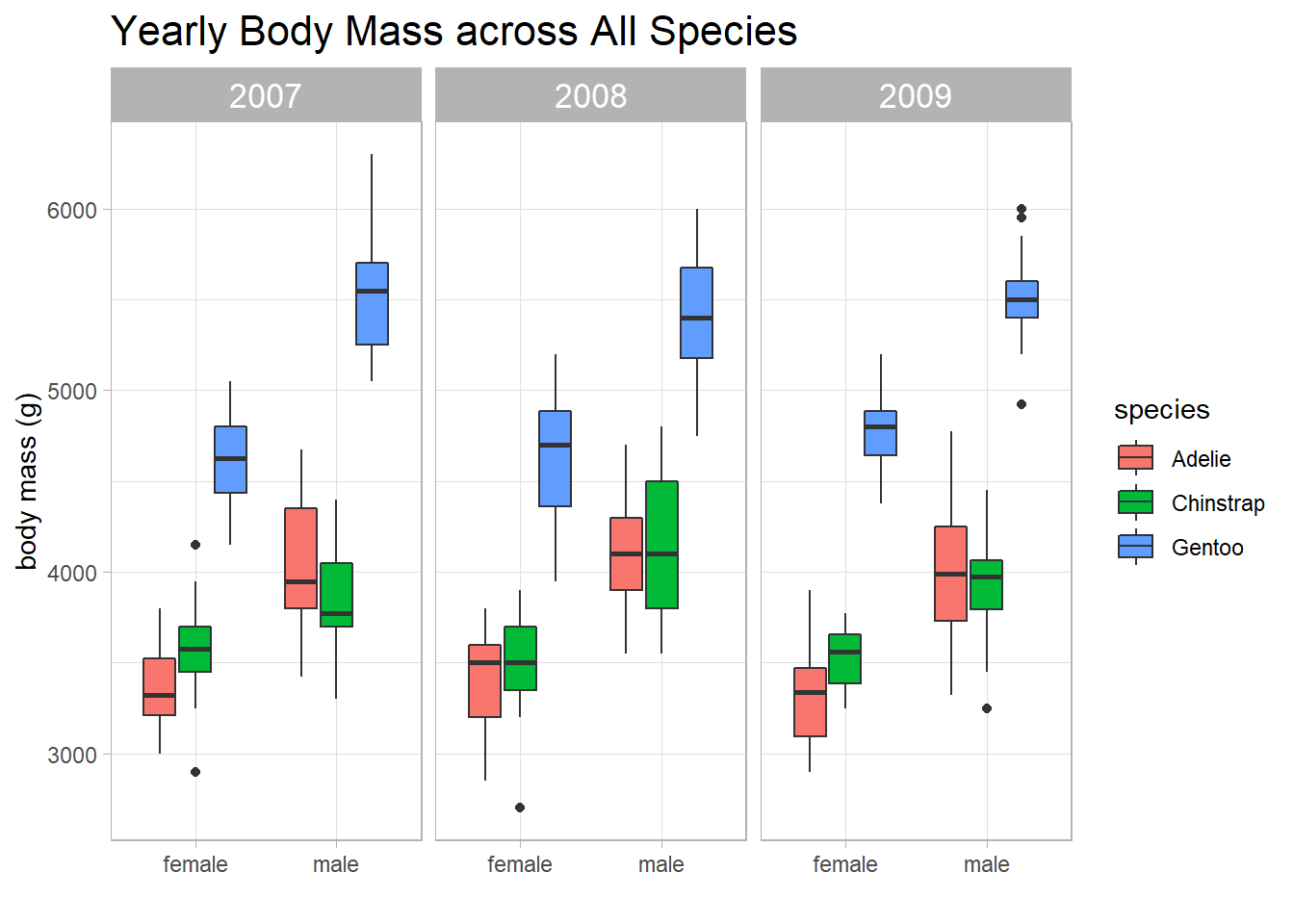
Body index distributions:
penguin_pivot <- penguins %>%
rename(`bill length (mm)` = bill_length_mm,
`bill depth (mm)` = bill_depth_mm,
`flipper length (mm)` = flipper_length_mm,
`body mass (g)` = body_mass_g) %>%
pivot_longer(c(3:6), names_to = "variable")
penguin_pivot %>%
ggplot(aes(value, fill = sex)) +
geom_histogram(alpha = 0.7) +
facet_wrap(~variable, scales = "free") +
labs(x = "respective body value",
fill = NULL,
title = "How Are Penguins' Body Indexes Distributed?") +
theme(strip.text = element_text(size = 13),
plot.title = element_text(size = 16))
Machine Learning:
I normally use the caret package to do my machine learning work, but now I am transferring to using tidymodels, as it does provide a coherent ecosystem as tidyverse provides. Since I am learning how to use tidymodels, this dataset is a great way to get more familiarity on how to use it. This machine learning section is inspired by Julia Silge's code and David Robinson's code. Many functions built inside the meta-package are my first time using them. Another note here is some code might be identical to the links provided above, with some slight modification.
I. Predicting the penguins' sex
#split up the data into training and testing data by using initial_split()
set.seed(2022)
penguin_split <- initial_split(penguins, prop = 0.8, strata = sex)
penguin_training <- training(penguin_split)
penguin_testing <- testing(penguin_split)
training_splits <- penguin_training %>%
vfold_cv(10)# logistic regression
glm_set <- logistic_reg() %>%
set_engine("glm")
# random forest classification
rf_set <- rand_forest() %>%
set_engine("ranger") %>%
set_mode("classification")# define a work flow
penguin_wf <- workflow() %>%
add_formula(factor(sex) ~ bill_length_mm + bill_depth_mm + flipper_length_mm + body_mass_g)
# add models here
glm_rs <- penguin_wf %>%
add_model(glm_set) %>%
fit_resamples(training_splits,
control = control_resamples(save_pred = TRUE))
rf_rs <- penguin_wf %>%
add_model(rf_set) %>%
fit_resamples(training_splits,
control = control_resamples(save_pred = TRUE))
collect_metrics(rf_rs)## # A tibble: 2 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.895 10 0.0157 Preprocessor1_Model1
## 2 roc_auc binary 0.951 10 0.0152 Preprocessor1_Model1Model evaluations:
collect_metrics(glm_rs)## # A tibble: 2 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.895 10 0.0155 Preprocessor1_Model1
## 2 roc_auc binary 0.955 10 0.0126 Preprocessor1_Model1collect_metrics(rf_rs)## # A tibble: 2 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.895 10 0.0157 Preprocessor1_Model1
## 2 roc_auc binary 0.951 10 0.0152 Preprocessor1_Model1It looks like the linear model has a slightly better fitting than its random forest counterpart.
Confusion Matrix:
glm_rs %>%
conf_mat_resampled()## # A tibble: 4 x 3
## Prediction Truth Freq
## <fct> <fct> <dbl>
## 1 female female 11.5
## 2 female male 1.1
## 3 male female 1.7
## 4 male male 12.3rf_rs %>%
conf_mat_resampled()## # A tibble: 4 x 3
## Prediction Truth Freq
## <fct> <fct> <dbl>
## 1 female female 11.8
## 2 female male 1.4
## 3 male female 1.4
## 4 male male 12glm_rs %>%
collect_predictions() %>%
rename(sex = `factor(sex)`) %>%
group_by(id) %>%
roc_curve(sex, .pred_female) %>%
ggplot(aes(1 - specificity, sensitivity, color = id)) +
geom_abline(lty = 2, size = 1.5) +
geom_path(show.legend = FALSE, alpha = 0.6, size = 1) +
coord_fixed(ratio = 1) +
labs(title = "The ROC Curves for All 10 Folds")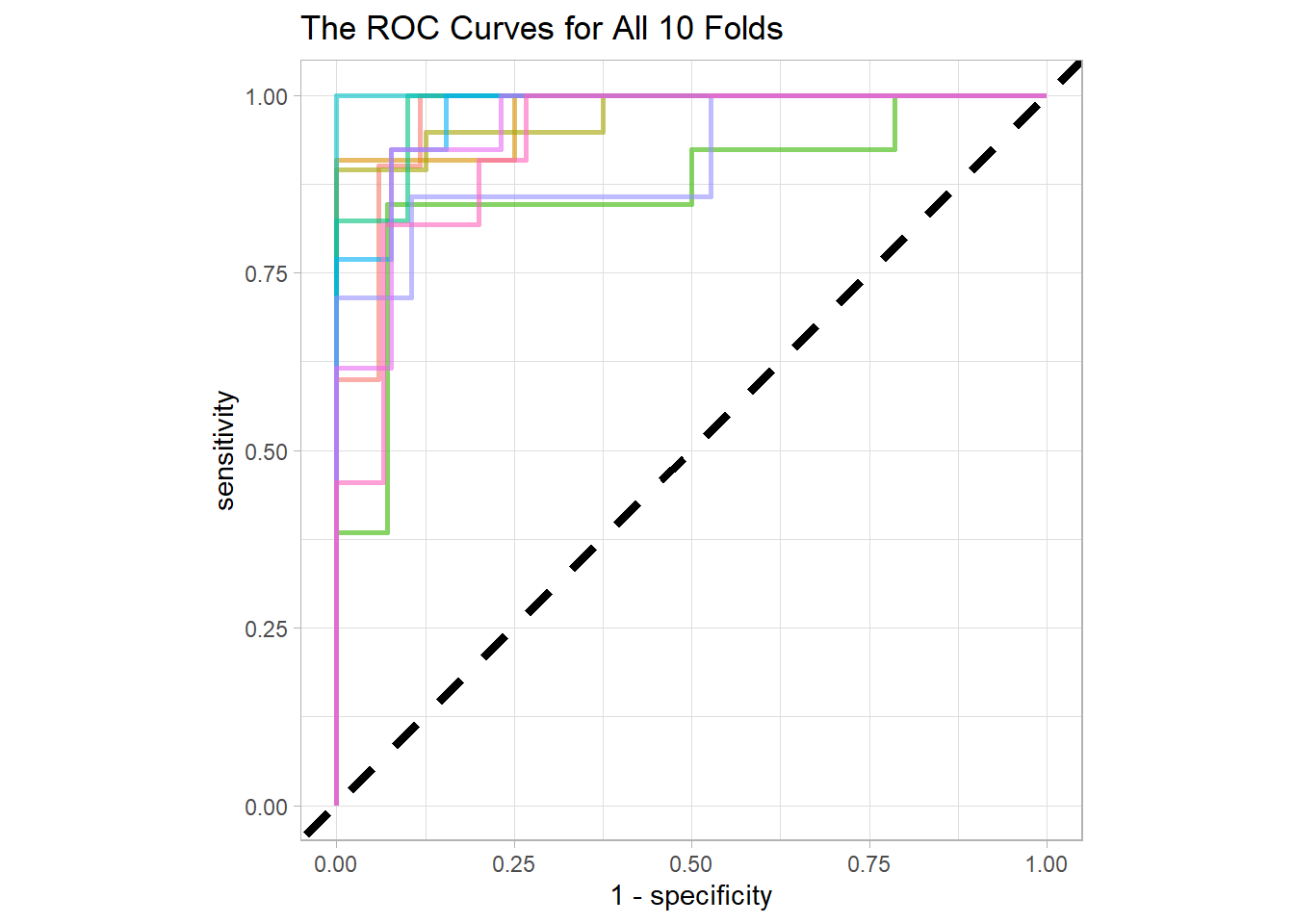
Final Model
penguin_final <- penguin_wf %>%
add_model(glm_set) %>%
last_fit(penguin_split)
penguin_final %>%
unnest(.predictions) %>%
rename(sex = 9) %>%
conf_mat(sex, .pred_class) ## Truth
## Prediction female male
## female 31 3
## male 2 31penguin_final$.workflow[[1]] %>%
tidy(exponentiate = T) ## # A tibble: 5 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 1.81e-24 9.06 -6.03 1.62e- 9
## 2 bill_length_mm 1.12e+ 0 0.0514 2.14 3.21e- 2
## 3 bill_depth_mm 7.27e+ 0 0.265 7.48 7.62e-14
## 4 flipper_length_mm 9.69e- 1 0.0389 -0.809 4.18e- 1
## 5 body_mass_g 1.01e+ 0 0.000903 5.87 4.43e- 9II. Predicting penguins' species
penguin_wf2 <- workflow() %>%
add_formula(species ~ bill_length_mm + bill_depth_mm + flipper_length_mm + body_mass_g)species_model <- penguin_wf2 %>%
add_model(rf_set) %>%
fit_resamples(training_splits,
metrics = metric_set(accuracy, kap, roc_auc),
control = control_resamples(save_pred = TRUE))
species_model %>%
collect_metrics()## # A tibble: 3 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy multiclass 0.970 10 0.0121 Preprocessor1_Model1
## 2 kap multiclass 0.954 10 0.0184 Preprocessor1_Model1
## 3 roc_auc hand_till 0.999 10 0.00116 Preprocessor1_Model1penguin_wf2 %>%
add_model(rf_set) %>%
last_fit(penguin_split) %>%
unnest(.predictions) %>%
conf_mat(species, .pred_class) ## Truth
## Prediction Adelie Chinstrap Gentoo
## Adelie 29 2 0
## Chinstrap 1 10 0
## Gentoo 0 0 25All models above perform fantastically well, in part due to the small size of the dataset and the easy prediction.
Working on penguins_raw
penguins_raw <- penguins_raw %>%
rename(delta_15 = delta_15_n_o_oo,
delta_13 = delta_13_c_o_oo)
penguins_raw %>%
count(delta_13, sort = T)## # A tibble: 332 x 2
## delta_13 n
## <dbl> <int>
## 1 NA 13
## 2 -27.0 1
## 3 -27.0 1
## 4 -26.9 1
## 5 -26.9 1
## 6 -26.9 1
## 7 -26.9 1
## 8 -26.8 1
## 9 -26.8 1
## 10 -26.8 1
## # ... with 322 more rowspenguins_raw %>%
ggplot(aes(delta_13, delta_15, color = sex, size = body_mass_g, shape = species)) +
geom_point(alpha = 0.8) +
labs(x = "Delta 13",
y = "Delta 15",
size = "body mass (g)",
title = "The Relations betweeen delta 13 and 15 across All Species and Sexes",
subtitle = "Delta 13 and 15 are the measurements of dietary comparison") +
theme(plot.title = element_text(size = 16))
The heavier of penguins are, the less Delta 15 and 13 their measurements are for both male and female penguins for Adelie and Gentoo species in particular.
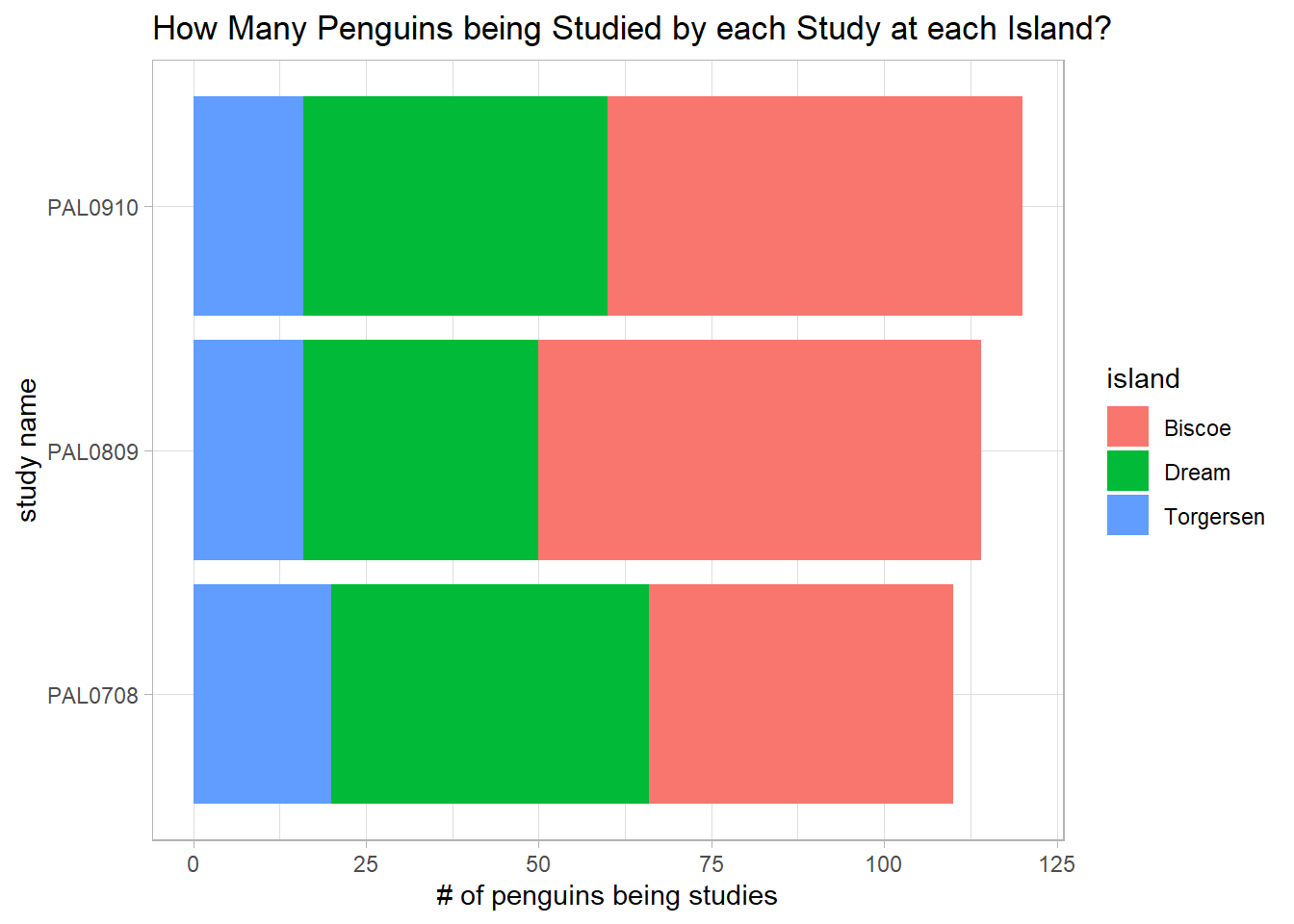
penguins_raw %>%
count(study_name, island, sort = T) %>%
ggplot(aes(n, study_name, fill = island)) +
geom_col() +
labs(x = "# of penguins being studies",
y = "study name",
title = "How Many Penguins being Studied by each Study at each Island?")